|
Hi! I am a third year CSE PhD student at University of Washington in the AIMS Lab, advised by Dr. Su-In Lee. Prior to starting my Ph.D, I spent a year as a data engineer at Microsoft in the Windows Experience team. I completed my Masters from Stanford in Computer Science with a depth in AI, where I was a research asistant in the Computational Neuroimage Science Lab (CNSLAB), advised by Dr. Kilian Pohl. I was also part of the AI for Healthcare (AIHC) bootcamp in the Stanford ML Group, advised by Dr. Pranav Rajpurkar and Dr. Andrew Ng. I finished my Bachelor's at Georgia Institute of Technology, with a major in Computer Engineering and a minor in Computer Science. I am also a part-time instructor at Persolv, teaching AI fundamentals to high school students. In my spare time, I like playing tennis, hiking, exploring different cuisines, and watching movies (especially legal thrillers). |

|
|
The research problems I want to work on lie at the interesection of Artificial Intelligence and Healthcare. Some of my current research interests include:
|
|
|
|
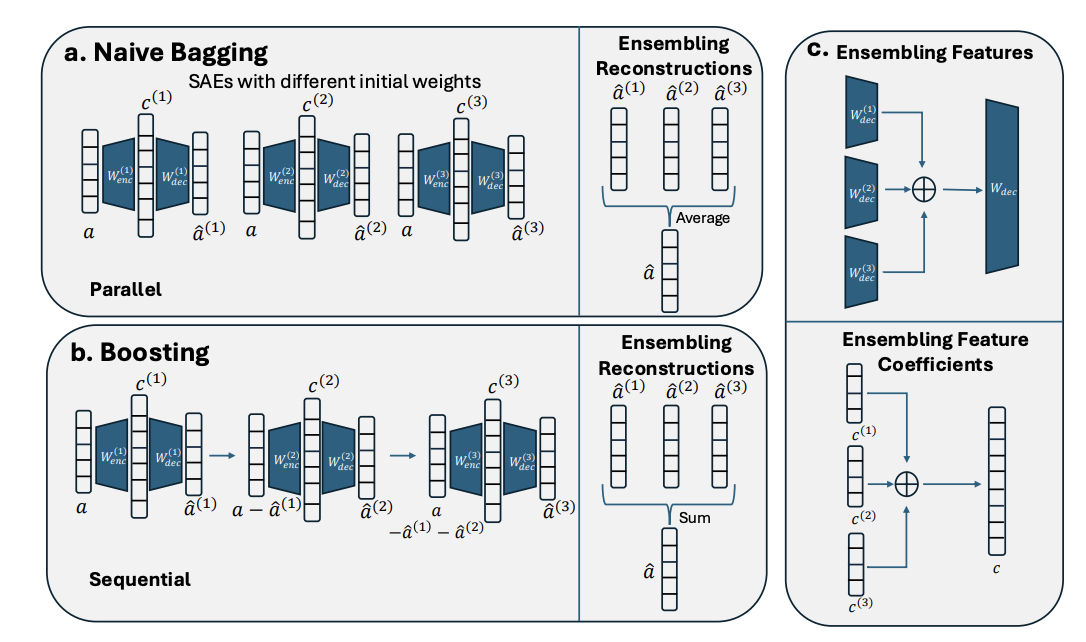
|
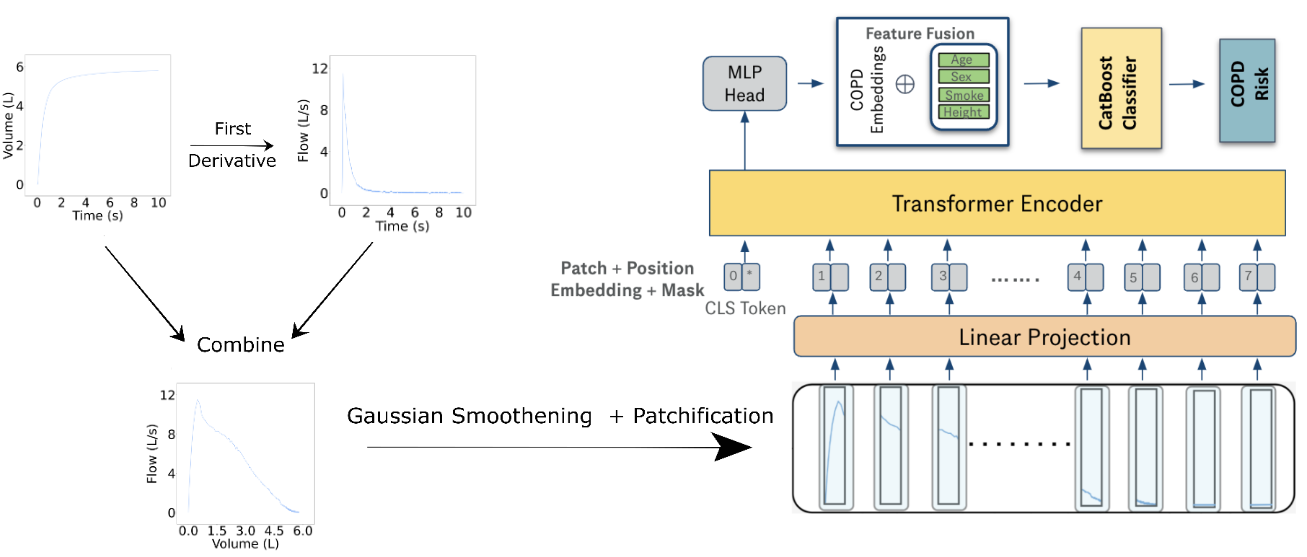
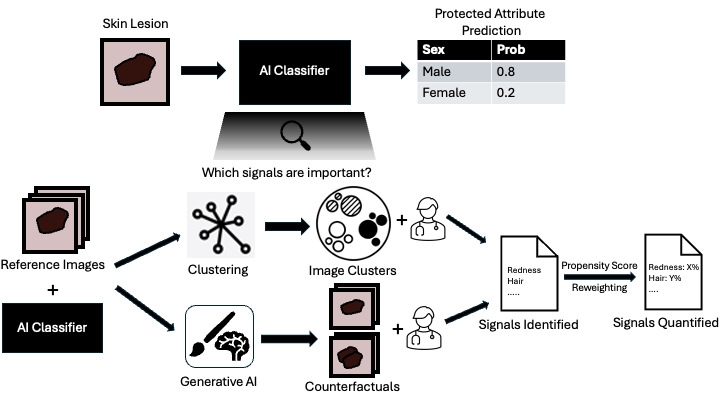
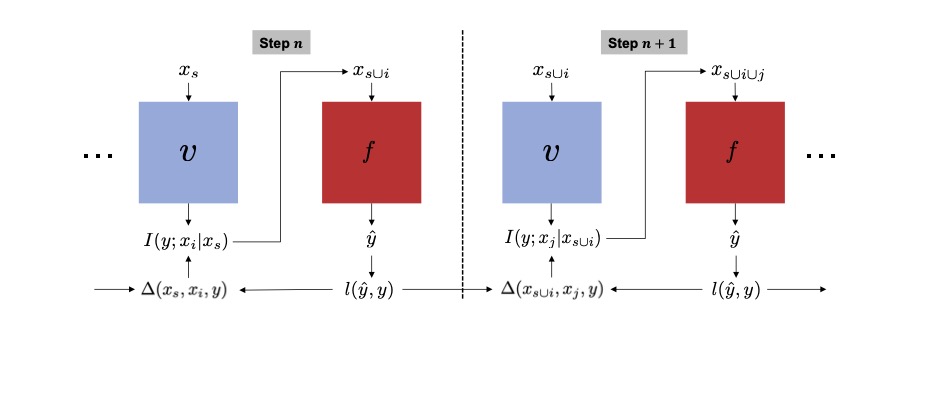
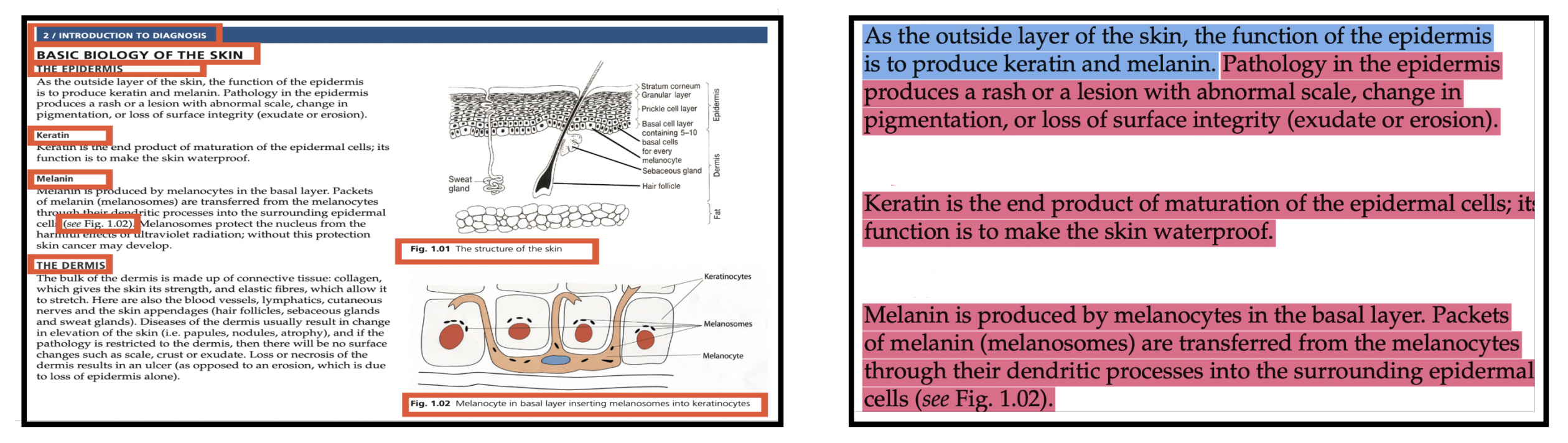
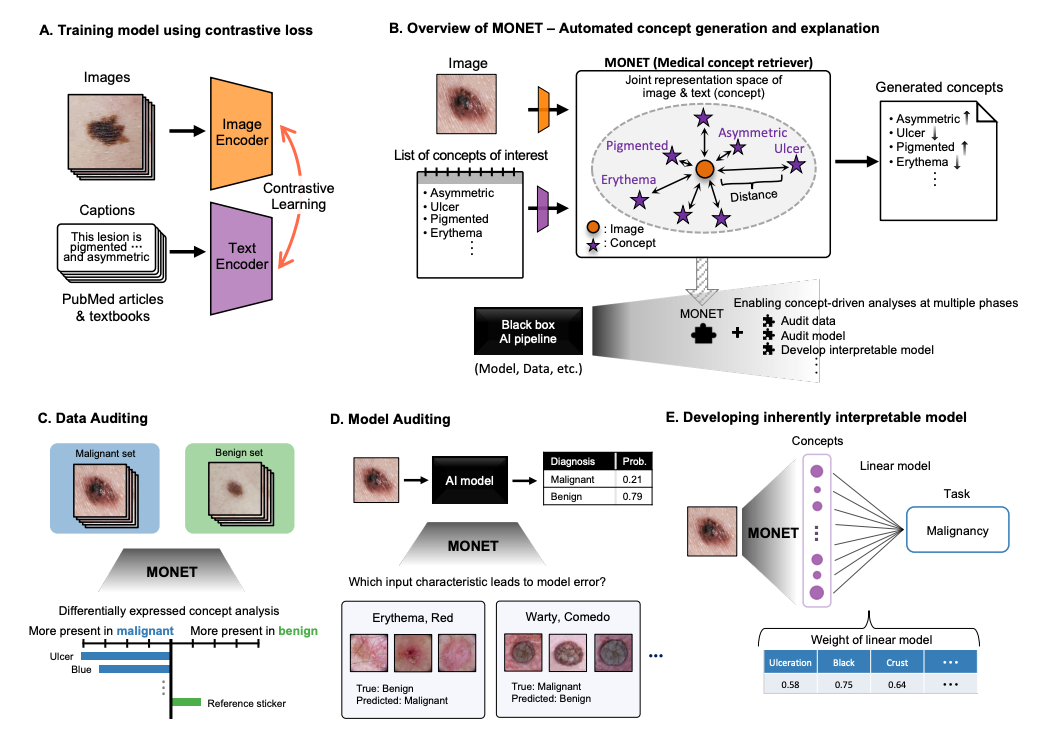
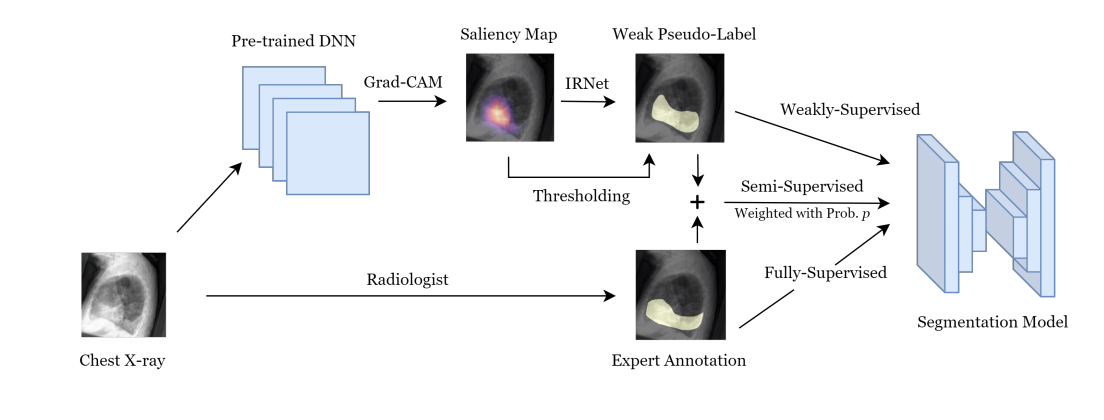
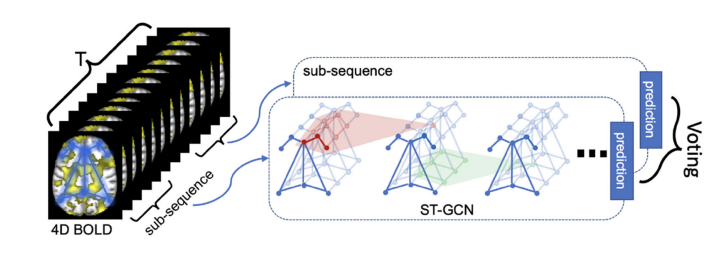
Soham Gadgil*, Chris Lin*, Su-In Lee Preprint (Under Review) [Paper] |
|
|
|
|
|
University of Washington |

|
The course covers principal ideas and developments in artificial intelligence: Problem solving and search, game playing, knowledge representation and reasoning, uncertainty, machine learning, natural language processing. I held weekly office hours and assisted in preparing/grading the homework assignments. |
Stanford |

|
TA for CS 107, one of the largest introductory undergraduate courses at Stanford with over 150 students. I led two hour-long lab sessions each week along with office hours and assisted the professor in grading homework and desiging exams. Topics included the C programming language, data representation, machine-level code, computer arithmetic, elements of code compilation, optimization of memory and runtime performance, and memory organization and management. |
|
TA for the first course offering of CS 329T. I co-developed and led the lab sections with ~25 students. I also helped the instructors design some of the lecture slides, homework assignments, and the final project. |
Georgia Tech |
|
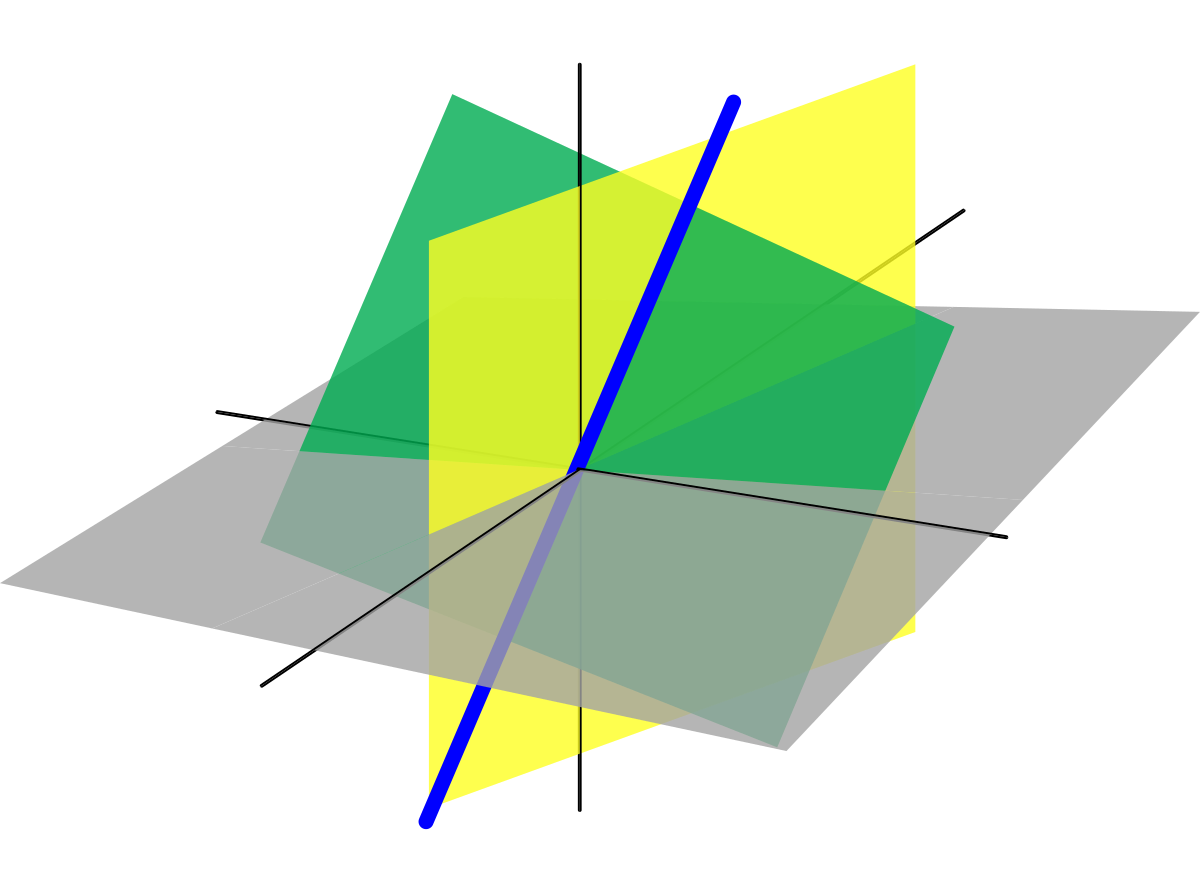
As a TA for linear algebra, I led two 50 minute recitation sessions with 25 students each week. Concepts ranged from eigenvalues, eigenvectors, applications to linear systems, least squares, diagonalization, quadratic forms. |
|
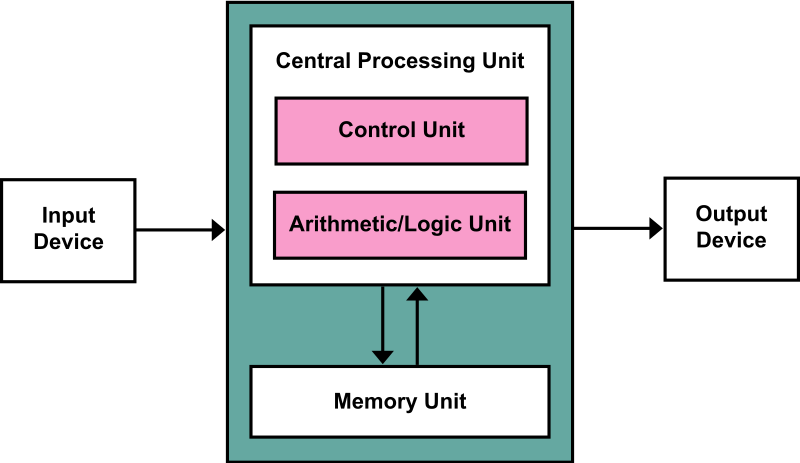
Guided over 60 students with homeworks and projects in computer architecture. Held weekly office hours, exam review sessions, and collaborated with the instructor for grading and project ideation. Topics included the basic organizational principles of the major components of a processor - the core, memory hierarchy, and the I/O subsystem. |
|
(Design and CSS courtesy: Jon Barron and Amlaan Bhoi) |